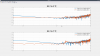
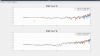
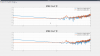
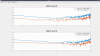
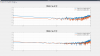
Last week we took advantage of the down time to do Z diagonalization measurements on the BSC ISI's, as Jeff outlined in his alog 36360. I used the data we got to calculate and install coefficients on all of the chambers that needed decoupling. The first five plots are the closeout Z to X/Y transfer functions comparing the before and after results showing the reduced coupling via tilt from Z drive to X/Y motion seen by the St1 T240s. For these plots, blue is always before, red is the after measurement. I forgot to include Y units but they are in nm/s / nm*rthz because I didn't convert the T240 signal to displacement.
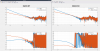
Yesterday I looked at repeating this on the output HAM's, I was just efficient enough to get HAM4 mostly done, but I need more time finish HAMs 2,3,5,6. The last attached plot is the Z to X/Y plots before and after for HAM4. The magnitude plots are in nm / nm*rthz. I also got data for HAM4 Y to RX, but it looks like the ISI is good enough in that DOF. HAM5 was similarly clean for Z to RX/RY, and I have a coefficient calculated for HAM6 Z to RX, but I would like to do a measurement after installing the cpsalign elements before I make a permanent change.
The measurements for the HAMs are essentially the same as the BSCs, driving at the inputs to isolation banks while the ISI is isolated with high blends (750mhz for the HAMs) with sensor correction off. Drives are usually 1-5e7 in awggui, for as long as you have patience for.
For the record, and to be explicit for future reference, here're the BSC-ISI ST1 tilt decoupling matrix elements (H1:ISI-${CHAMBER}_ST1_CPSALIGN_${M}_${N}) that Jim has now installed. One reads the matrix as "The corrected RX and RY CPS signals used in the isolation loops contain small portions of X, Y, and Z signals."
ETMX misaligned
X Y Z
a
l X 1 0 0
i Y 0 1 0
g Z 0 0 1
n RX -0.0030 -0.0020 +0.0060
e RY +0.0013 +0.0001 -0.0025
d RZ 0 0 0
ETMY misaligned
X Y Z
a
l X 1 0 0
i Y 0 1 0
g Z 0 0 1
n RX +0.0010 -0.0010 0
e RY +0.0005 +0.00075 0
d RZ 0 0 0
ITMX misaligned
X Y Z
a
l X 1 0 0
i Y 0 1 0
g Z 0 0 1
n RX +0.0015 -0.0025 0
e RY +0.0015 +0.0015 +0.0025
d RZ 0 0 0
ITMY misaligned
X Y Z
a
l X 1 0 0
i Y 0 1 0
g Z 0 0 1
n RX +0.0015 -0.0007 -0.0057
e RY +0.00075 +0.0005 +0.0030
d RZ 0 0 0
BS misaligned
X Y Z
a
l X 1 0 0
i Y 0 1 0
g Z 0 0 1
n RX -0.0001 -0.0007 -0.0035
e RY +0.00075 0 +0.0025
d RZ 0 0 0